The Jpeg DNA algorithm¶
The Jpeg DNA python package is an implementation of the algorithm of the same name. The main purpose of the Jpeg DNA algorithm is to encode the DCT coefficients into DNA-like quaternary code instead of binary code.
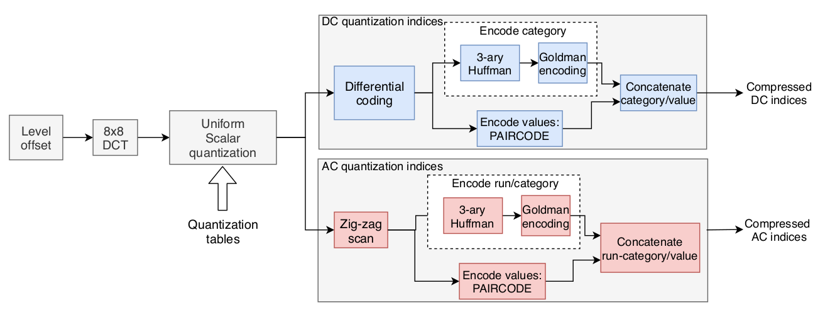
The objective of the encoder is to transform the matrix of DCT coefficients into a stream of quaternary data. First, a quantization is applied, then a zigzag transform, the result is a sequence of integers in which the first element is the DC coefficient, then follow all the 63 AC coefficients. The encoding system for DC and for AC coefficients share similarities but have heir own specificity. We will first consider in depth the case of the DC values. Firstly, the DC coefficient is differentially coded according to the previous DC coefficient (the DC coefficient of the previous block if it exists, otherwise 0). The result of this differential coding is then given a category in function of its absolute value. The higher the absolute value, the higher the category. This category ordering system will find its importance when explaining the way we encode the values. There is a finite number of categories (less than ten). We encode those categories using a ternary Huffman Tree coder. The result of this operation is a codebook of codewords composed of 0,1 and 2. To convert this to a DNA like data, we use a Goldman coder with {A, C, G, T} as alphabet. To make this is possible, we need to be able to create the Huffman Tree associated with those categories. In other words, we need to know the frequency appearances of each category. This can either be precomputed from the image before starting the encoding, or it is possible to us more generalist frequency tables computed from a lot of images. That last choice will be sub-efficient in terms of accuracy of those frequencies, but no frequency tables will be transmitted to the decoder, we will expect it to already possess those generalist frequency tables. For the moment, we only have explained the encoding of the category of the coefficient. As explained before and as can be shown in the table above, this category gives us an interval of values in which the coefficient falls. The smaller the category, the smaller the interval. For each interval, meaning for each category, a fixed-length codebook will be used. The smaller the codebook, the smaller the codewords that it contains. We then use a simple bijection between the possible values and the codebook indices to encode the values with a unique element of the codebook. The final result of encoding a DC differential coefficient is the concatenation of the codeword for its category and of the codeword for its value. During decoding, using Huffman, we will first decode the category, deduce the value codebook to use, obtain the index of the value codeword in the fixed-length codebook, use the inverse of the bijection to get the value, and then inverse the differential coding. The way AC coefficients are encoded is quite similar as it can be seen in the block schema at the beginning of the section. We still use a system of category, and we still use a fixed-length codebook to encode the values according to their category, but the Huffman/Goldman coders don’t encode the category anymore. They now encode a run/category which describes with a focus on zeros a pattern met in the zigzag sequence of AC coefficients. Two of the possible patterns are not associated with a value and don’t follow the system of category and value previously described. Those patterns are a sequence of 16 zeros, and an end of sequence only composed of zeros. When the sequence is read, the number of zeros is counted, up to 16. Every time it gets to 16, the 16 zeros run/category codeword is generated and the counter of zeros is reinitialized to 0. When a non-zero coefficient is met, the run/category coder will encode a coupling of values: the number of zeros preceding this value (up to 15) and the category of the current coefficient, as described in the DC coefficient encoding process. We then encode the value with the same codebook system than for the DC values and concatenate the result of both run/category and value coders. We concatenate all the DC coefficient code with all the run/category/values codes to obtain a stream for the block. We concatenate the block codes to obtain the stream for the image.
The development of this algorithm has been oriented towards an easy-to-use, easy-to-modify paradigm. As previously described, the algorithm is working with a collection of coders (Huffman, Goldman), transforms (DCT, Zigzag). We have decided to make it as easy as possible for someone to change one part of the algorithm, for instance a coder, without having to change everything else. The library is composed of three main packages, one for coders, one for transforms and one for codecs. The latter one only contains the Jpeg DNA codec for the moment but anyone that would want to create a new codec would implement it here. The coders and transforms used by these codecs will be developed in the packages of the same name. Then on a list package called scripts, will find and maybe develop general scripts for those codecs, for compression, decompression and performance evaluation.